Hossein Kashfi
Generating and Utilizing Murine Cas9-Expressing Intestinal Organoids for Large-Scale Knockout Genetic Screening
Kashfi, Hossein; Jinks, Nicholas; Nateri, Abdolrahman S.
Abstract
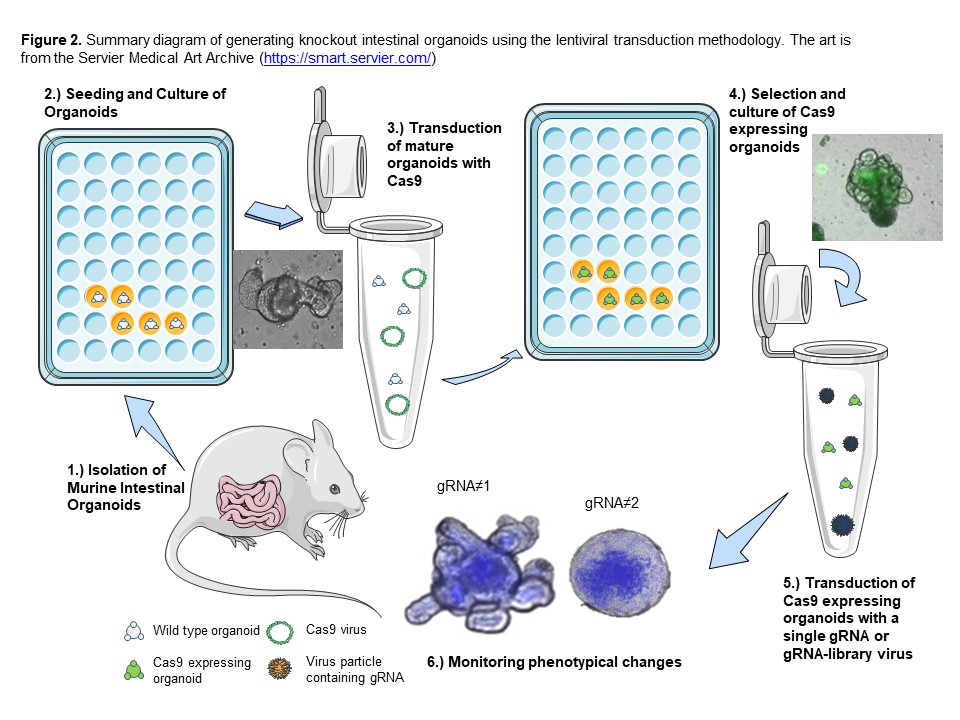
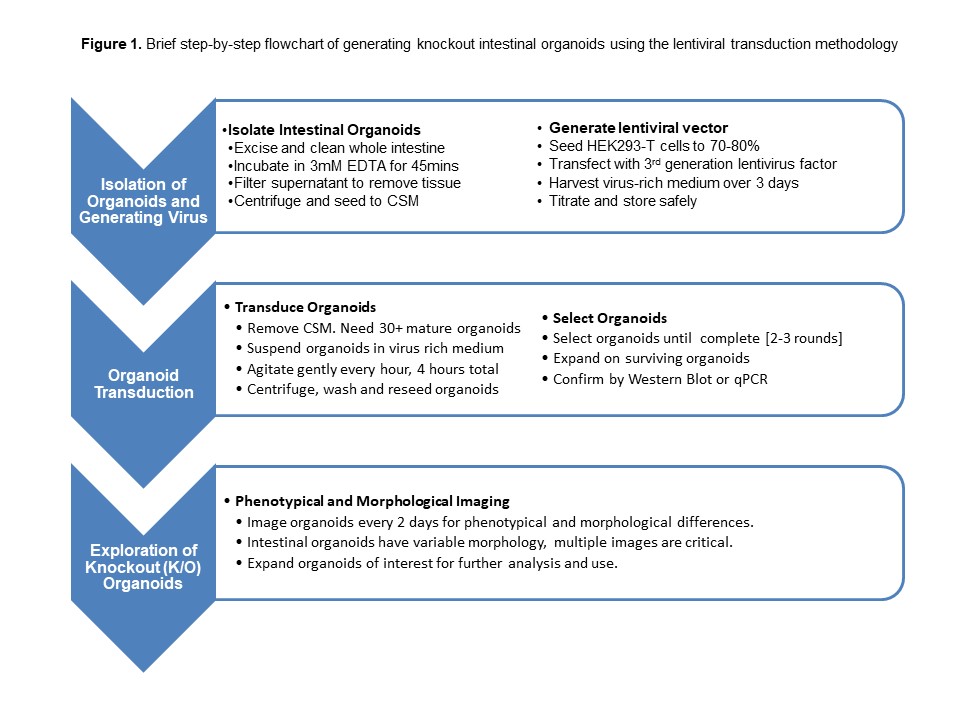
Organoid culture faithfully reproduces the in vivo characteristics of the intestinal/colon epithelium and elucidates molecular mechanisms underlying the regulation of stem cell compartment that, if altered, may lead tumorigenesis. CRISPR-Cas9 based editing technology has provided promising opportunities for targeted loss-of-function mutations at chosen sites in the genome of eukaryotes. Herein, we demonstrate a CRISPR/Cas9-mediated mutagenesis-based screening method using murine intestinal organoids by investigating the phenotypical morphology of Cas9-expressing murine intestinal organoids. Murine intestinal crypts can be isolated and seeded into Matrigel and grown into stable organoid lines. Organoids subsequently transduced and selected to generate Cas9 expressing organoids. These organoids can be further transduced with the second lentiviruses expressing guide RNA (gRNA) (s) and screened for 8–10 days using bright-field and fluorescent microscopy to determine possible morphological or phenotypical abnormalities. Via phenotypical screening analysis, the candidate knockouts can be selected based on differential abnormal growth pattern vs their untransduced or lenti-GFP transduced controls. Further assessment of these knockout organoids can be done via phalloidin and propidium iodide (PI) staining, proliferation assay and qRT-PCR and also biochemical analysis. This CRISPR/Cas9 organoid mutagenesis-based screening method provides a reliable and rapid approach for investigating large numbers of genes with unknown/poorly identified biological functions. Knockout intestinal organoids can be associated with the key biological function of the gene(s) in development, homeostasis, disease progression, tumorigenesis, and drug screening, thereby reducing and potentially replacing animal models.
Citation
Kashfi, H., Jinks, N., & Nateri, A. S. (2020). Generating and Utilizing Murine Cas9-Expressing Intestinal Organoids for Large-Scale Knockout Genetic Screening. Methods in Molecular Biology, 2171, 257-269. https://doi.org/10.1007/978-1-0716-0747-3_17
| Journal Article Type | Article |
|---|---|
| Acceptance Date | Jun 30, 2020 |
| Online Publication Date | Jul 24, 2020 |
| Publication Date | 2020 |
| Deposit Date | Sep 16, 2020 |
| Publicly Available Date | Jul 25, 2021 |
| Journal | Methods in Molecular Biology |
| Peer Reviewed | Peer Reviewed |
| Volume | 2171 |
| Pages | 257-269 |
| DOI | https://doi.org/10.1007/978-1-0716-0747-3_17 |
| Public URL | https://nottingham-repository.worktribe.com/output/4795956 |
| Publisher URL | https://link.springer.com/protocol/10.1007%2F978-1-0716-0747-3_17#aboutcontent |
| Additional Information | 9781071607466 This is a post-peer-review, pre-copyedit version of an article published in Methods in Molecular Biology. The final authenticated version is available online at: https://doi.org/10.1007/978-1-0716-0747-3_17. Kashfi H., Jinks N., Nateri A.S. (2020) Generating and Utilizing Murine Cas9-Expressing Intestinal Organoids for Large-Scale Knockout Genetic Screening. In: Ordóñez-Morán P. (eds) Intestinal Stem Cells. Methods in Molecular Biology, vol 2171. Humana, New York, NY. |
Files
Figure 2
(156 Kb)
Image
Figure 1
(131 Kb)
Image
Final Kashfi Jinks Et Al. Cas9- Organoid Method R
(265 Kb)
PDF
You might also like
Mast Cell Tryptase Release Contributes to Disease Progression in Lymphangioleiomyomatosis
(2021)
Journal Article
FLYWCH1, a Multi-Functional Zinc Finger Protein Contributes to the DNA Repair Pathway
(2021)
Journal Article
Downloadable Citations
About Repository@Nottingham
Administrator e-mail: discovery-access-systems@nottingham.ac.uk
This application uses the following open-source libraries:
SheetJS Community Edition
Apache License Version 2.0 (http://www.apache.org/licenses/)
PDF.js
Apache License Version 2.0 (http://www.apache.org/licenses/)
Font Awesome
SIL OFL 1.1 (http://scripts.sil.org/OFL)
MIT License (http://opensource.org/licenses/mit-license.html)
CC BY 3.0 ( http://creativecommons.org/licenses/by/3.0/)
Powered by Worktribe © 2025
Advanced Search